Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
This page serves as documentation for navigating queries in the SPARC Connectivity Knowledge base of the Autonomic Nervous system (SCKAN).
Background information about SCKAN can be viewed here. Documentation for Setup, installation, queries and tutorials are accessible via this link. SCKAN contains knowledge about Central nervous system-Autonomic nervous system-end organ circuitry derived from SPARC data and scientific literature, in a form that supports reasoning. This gives users the ability to query the knowledge in SCKAN, which was captured through interviews with SPARC investigators, anatomical experts and the scientific literature. The sources used to create this knowledge are located here. The section below provides example queries for one source of knowledge in SCKAN, ApiNATOMY. ApiNATOMY is a knowledge model and tool suite specifically created to represent biological circuit connectivity. A circuit represents a detailed model of ANS connectivity associated with a particular organ, e.g., bladder or functional circuits, e.g., defensive breathing. These represent neuron populations that give rise to ANS connections, and include mappings of the locations of cell bodies, dendrites, axon segments and synaptic endings involved in a particular circuit. The ApiNATOMY models in the current release of SCKAN are listed below.
- Individual Neuron Populations are labeled: 1 10 11 12 13 14 15 16 17 18 19 2 20 21 22 23 24 25 26 27 28 29 3 4 5 6 7 8 9
- Individual Neuron Populations are labeled: 1 10 11 12 13 14 15 16 17 18 19 2 20 3 4 5 6 7 8 9
- Individual Neuron Populations are labeled: 1 2 3 4 5 6
- Individual Neuron Populations are labeled: 1 10a 10v 11 12 13 2i 2m 4 5 6 7a 7v 8a 8v 9a 9v
- Individual Neuron Populations are labeled: b c d f g h i j j’ k l l’ m n o p q
- Individual Neuron Populations are labeled: 1 10 11 12 13 2 3 4 5 6 7 8 9
- Individual Neuron Populations are labeled: 1 2 3 4 5
- Individual Neuron Populations are labeled:
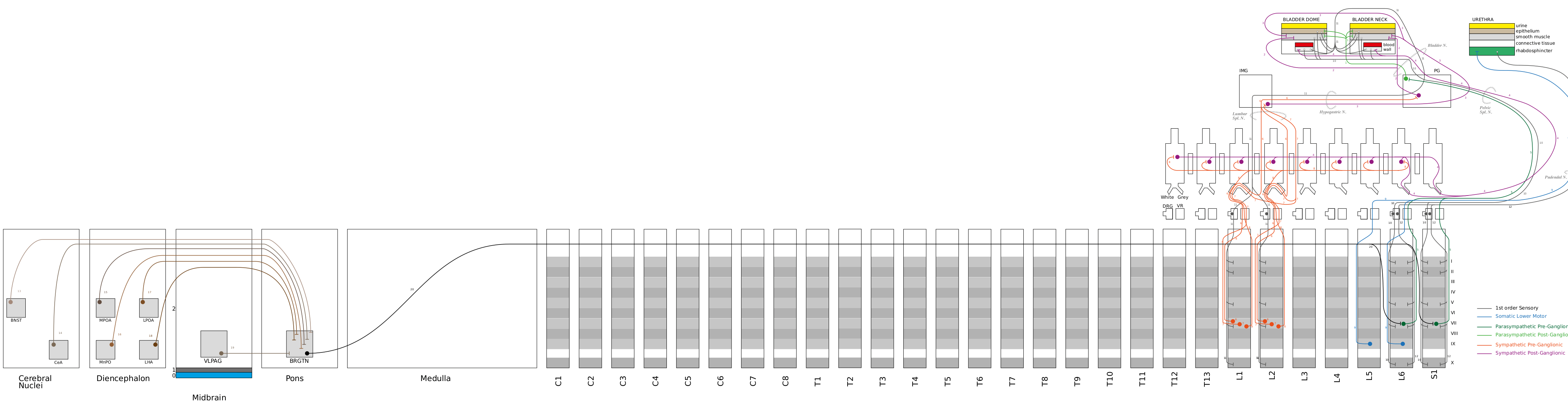
A graphical representation of the Keast ApiNATOMY model of bladder innervation (kblad) is shown below
SCKAN can be used to query circuit connection from ApiNATOMY models, individual connection from literature curation and natural language processing and datasets within SPARC, the below queries are simple examples to introduce the formalism of the models.
Please see this page for more information on how to access queries. Setup, installation, queries, and tutorial documentation is accessible via this link.
Example queries -
Two example queries are listed below. Please select the text to expand the section and view more information about the query.
Query 1 : Return all of the anatomical regions that neuron population 5 travels to in the Keast ApiNATOMY model (kblad)
Background information about this query:
This query will retrieve knowledge from SCKAN about the neural circuits (neuron population 5) in the Keast ApiNATOMY model of the bladder (kblad) innervating the urinary bladder and urethra. The neurons in this neuronal circuit, neuron population 5, refers to a population of ParaSympathetic PReganglionic (PSPR) neurons with somas in L6 and S1 spinal cord segments, with axon terminals in the pelvic ganglia. The notation “neuron population 5” or “5” is used for internal labeling.

Short names and abbreviations used in the model are
Keast ApiNATOMY model of bladder innervation = Keast ApiNATOMY model = kblad
Neuron population 5 = 5
ParaSympathetic PReganglionic neuron = PSPR
Pelvic Ganglia = PG
Pelvic splanchnic nerve = Pelvic Spl N
L5 = L5 spinal cord segment
L6 = L6 spinal cord segment
As always, additional in-depth information about the ApiNATOMY knowledge model and the models can be found here. Additional abbreviations used in the models are found here.
Query results:
The results from the query can be viewed
1. In a web interface, as dynamic tree rendering of neurons in an ApiNATOMY model. The url for this query is http://ontology.neuinfo.org/trees/sparc/dynamic/demos/apinat/neru-5/ilxtr:neuron-type-keast-5. Below is an image of this page and brief introduction of how to read the page.
_7f1de29e6da19d22.png)
The results of this query will be displayed as a tree format, where the root node (value) will show the initial query and the subsequent nodes (values) will provide the results to the query.
Expected results (see figure below):
The results from the query will populate in multiple views. But first, to understand these projections, please see the schematic of the Keast ApiNATOMY model (kblad) is directly below this text.

The trajectory of interest is highlighted in purple.
This neuron population have
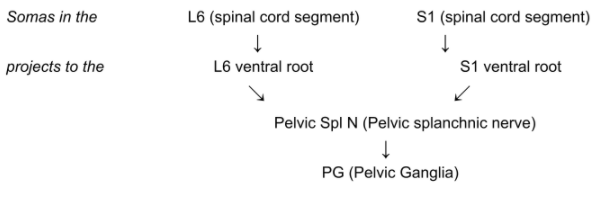
L6 (spinal cord segment)
L6 ventral root
S1 (spinal cord segment)
S1 ventral root
Pelvic Spl N (Pelvic splanchnic nerve)
- PG (Pelvic Ganglia)
Query 2 : Find all neurons, across all ApiNATOMY models, with processes in the Inferior Mesenteric Ganglion (IMG).
Background information about this query:
This query will retrieve knowledge from SCKAN to identify all neuron populations from all ApiNATOMY models with processes (an axon or dendrite) in the IMG. A list of the ApiNATOMY models in Release 1 are located below.
As with the example above, these processes will be identified from the neuron populations in the model. The notation “neuron population X” or “X” is used for internal labeling.
As always, additional in-depth information about the ApiNATOMY knowledge model and the models can be found here. Additional abbreviations used in the models are found here.
Query results:
The results from the query can only be viewed as a self-contained docker instance. More information about creating this instance is at this link.
Expected results (see figure below):
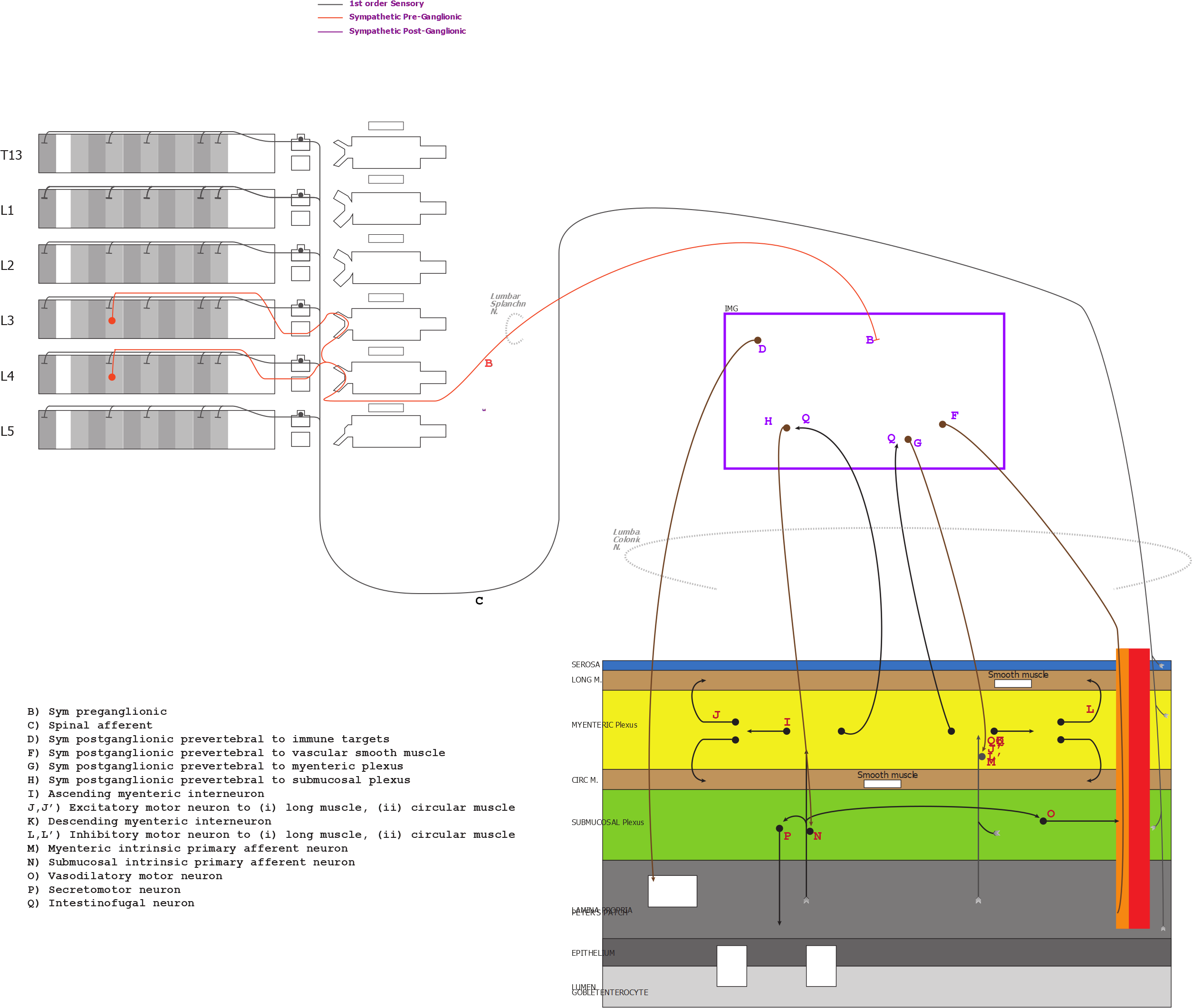
There are two APiNATOMY models with neuron processes in the IMG. The two models that fit this criteria and a schematic of these models are listed below. The areas of interest highlighted with purple.
1. Keast ApiNATOMY model of bladder innervation (kblad) - Neurons 3, 6, 7, 11

A list of all neurons are listed below by ApiNATOMY model
Keast ApiNATOMY model of bladder innervation (kblad)
Neuron 3
Soma to SPO neuron 3 in IMG_Neuron 3 (kblad)
Neuron 6
Soma of SPR neuron in L1_Neuron 6 (kblad)
Soma of SPR neuron in L2_Neuron 6 (kblad)
Neuron 7
Soma of SPR neuron in L1_Neuron 7 (kblad)
Soma of SPR neuron in L2_Neuron 7 (kblad)
Neuron 11
Soma of FOS in L1 DRG_Neuron 11 (kblad)
Soma of FOS in L2 DRG_Neuron 11 (kblad)
SAWG ApiNATOMY model of the distal colon (sdcol)
Neuron B
Soma of SPR neuron in L3_Neuron B (sdcol)
Soma of SPR neuron in L4_Neuron B (sdcol)
Neuron D
Soma of SPO neuron in IMG_Neuron D (sdcol)
Neuron F
Soma of SPO neuron in IMG_Neuron F (sdcol)
Neuron G
Soma of SPO neuron in IMG_Neuron G (sdcol)
Neuron H
Soma of SPO neuron in IMG_Neuron H (sdcol)
Neuron Q
- Soma of intestinofugal neuron in myenteric plexus of colon_Neuron Q (sdcol)
Short names and abbreviations used in the model are
- Keast ApiNATOMY model of bladder innervation = Keast ApiNATOMY model = kblad
- SAWG ApiNATOMY model of the distal colon (sdcol) = Distal Colon ApiNATOMY model = sdcol
- ParaSympathetic PReganglionic neuron = PSPR
- Sympathetic POstganglionic neuron = SPO
- Sympathetic PReganglionic neuron = SPR
- First order sensory neuron = FOS
- Neuron = neuron population in model
- Inferior Mesenteric Ganglion = IMG
- Dorsal Root Ganglion = DRG
- L1 spinal cord segment = L1
- L2 spinal cord segment = L2

