Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
Seek 'n Blastn
Seek n' Blastn' limitations are documented in the following manuscript:
Labbé, C., Cabanac, G., West, R.A. et al. Flagging incorrect nucleotide sequence reagents in biomedical papers: To what extent does the leading publication format impede automatic error detection?. Scientometrics 124, 1139–1156 (2020). https://doi.org/10.1007/s11192-020-03463-z
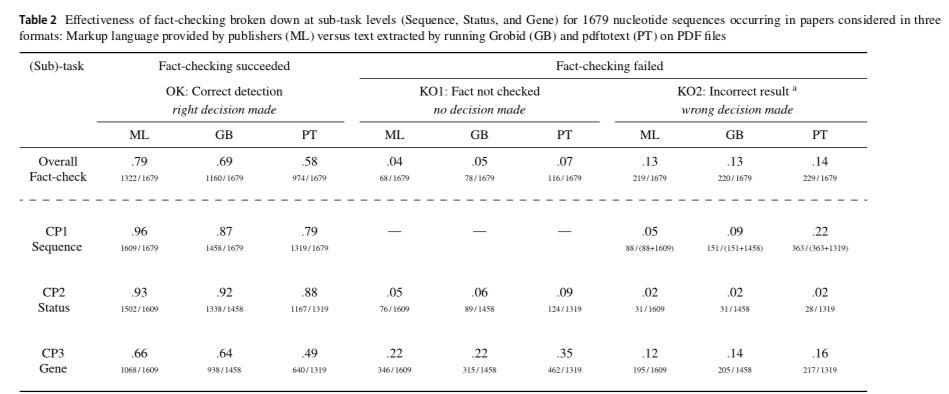
The contribution of this paper is twofold. First, we designed the erroneous reagent checking (ERC) benchmark to assess the accuracy of fact-checkers screening biomedical publications for dubious mentions of nucleotide sequence reagents. It comes with a test collection comprised of 1679 nucleotide sequence reagents that were curated by biomedical experts. Second, we benchmarked our own screening software called Seek&Blastn with three input formats to assess the extent of performance loss when operating on various publication formats. Our findings stress the superiority of markup formats (a 79% detection rate on XML and HTML) over the prominent PDF format (a 69% detection rate at most) regarding an error flagging task.
Briefly...